pygimli.physics.SIP#
Spectral induced polarization (SIP) measurements and fittings.
Overview#
Functions
|
|
|
|
|
|
|
|
|
|
|
|
|
Show amplitude spectrum (resistivity as a function of f). |
|
Show phase spectrum (-phi as a function of f). |
|
Shortcut to load SIP spectral data. |
|
Original complex-valued permittivity formulation (Cole&Cole, 1941). |
|
Frequency-domain Cole-Cole impedance model after Pelton et al. (1978). |
|
Frequency-domain double Cole-Cole resistivity (impedance) model. |
|
Complex-valued conductivity (admittance) Cole-Cole model. |
|
Complex-valued double added conductivity (admittance) model. |
|
For backward compatibility. |
|
Show amplitude and phase spectra in two subplots. |
|
Convert \(\tau_{\rho}\) to \(\tau_{\sigma}\) Cole-Cole model. |
Classes
|
Cole-Cole model with EM term after Pelton et al. (1978). |
|
Cole-Cole model with EM term after Pelton et al. (1978). |
|
Cole-Cole model with EM term after Pelton et al. (1978). |
|
Cole-Cole model with EM term after Pelton et al. (1978). |
|
Debye decomposition (smooth Debye relaxations) of complex data. |
|
Debye decomposition (smooth Debye relaxations) phase only. |
|
Double Cole-Cole model after Pelton et al. (1978). |
|
Cole-Cole model with EM term after Pelton et al. (1978). |
|
SIP spectrum data analysis. |
|
Manager to work with spectra data. |
|
Modelling framework with an array of freqencies as data space. |
Functions#
- pygimli.physics.SIP.drawAmplitudeSpectrum(ax, freq, amp, ylabel='$\\rho$ ($\\Omega$m)', grid=True, marker='+', ylog=True, **kwargs)[source]#
Show amplitude spectrum (resistivity as a function of f).
- pygimli.physics.SIP.drawPhaseSpectrum(ax, freq, phi, ylabel='$-\\phi$ (mrad)', grid=True, marker='+', ylog=False, **kwargs)[source]#
Show phase spectrum (-phi as a function of f).
- pygimli.physics.SIP.load(fileName, verbose=False, **kwargs)[source]#
Shortcut to load SIP spectral data.
Import Data and try to assume the file format.
- Parameters:
fileName (str)
- Returns:
freqs, amp, phi – Frequencies, amplitudes and phases phi in neg. radiant
- Return type:
np.array
- pygimli.physics.SIP.modelColeColeEpsilon(f, e0, eInf, tau, alpha)[source]#
Original complex-valued permittivity formulation (Cole&Cole, 1941).
- pygimli.physics.SIP.modelColeColeRho(f, rho, m, tau, c, a=1)[source]#
Frequency-domain Cole-Cole impedance model after Pelton et al. (1978).
Frequency-domain Cole-Cole impedance model after Pelton et al. (1978) [Pelton et al., 1978]
\[\begin{split}Z(\omega) & = \rho_0\left[1 - m \left(1 - \frac{1}{1+(\text{i}\omega\tau)^c}\right)\right] \\ \quad \text{with}\quad m & = \frac{1}{1+\frac{\rho_0}{\rho_1}} \quad \text{and}\quad \omega =2\pi f\end{split}\]\(Z(\omega)\) - Complex impedance per 1A current injection
\(f\) - Frequency
\(\rho_0\) – Background resistivity states the unblocked pore path
\(\rho_1\) – Resistance of the solution in the blocked pore passages
\(m\) – Chargeability after Seigel (1959) [Seigel, 1959] as being the ratio of voltage immediately after, to the voltage immediately before cessation of an infinitely long charging current.
\(\tau\) – ‘Time constant’ relaxation time [s] for 1/e decay
\(c\) - Rate of charge accumulation. Cole-Cole exponent typically [0.1 .. 0.6]
Examples
>>> import numpy as np >>> import pygimli as pg >>> from pygimli.physics.SIP import modelColeColeRho >>> f = np.logspace(-2, 5, 100) >>> m = np.linspace(0.1, 0.9, 5) >>> tau = 0.01 >>> fImMin = 1/(tau*2*np.pi) >>> fig, axs = pg.plt.subplots(1, 2) >>> ax1 = axs[0] >>> ax2 = axs[0].twinx() >>> ax3 = axs[1] >>> ax4 = axs[1].twinx() >>> for i in range(len(m)): ... Z = modelColeColeRho(f, rho=1, m=m[i], tau=tau, c=0.5) ... _= ax1.loglog(f, np.abs(Z), color='black') ... _= ax2.loglog(f, -np.angle(Z)*1000, color='b') ... _= ax3.loglog(f, Z.real, color='g') ... _= ax4.semilogx(f, Z.imag, color='r') ... _= ax4.plot([fImMin, fImMin], [-0.2, 0.], color='r') >>> _ = ax4.text(fImMin, -0.1, r"$f($min($Z''$))=$\frac{1}{2*\pi\tau}$", color='r') >>> _ = ax4.text(0.1, -0.17, r"$f($min[$Z''$])=$\frac{1}{2\pi\tau}$", color='r') >>> _ = ax1.set_ylabel('Amplitude $|Z(f)|$', color='black') >>> _ = ax1.set_xlabel('Frequency $f$ [Hz]') >>> _ = ax1.set_ylim(1e-2, 1) >>> _ = ax2.set_ylabel(r'- Phase $\varphi$ [mrad]', color='b') >>> _ = ax2.set_ylim(1, 1e3) >>> _ = ax3.set_ylabel('re $Z(f)$', color='g') >>> _ = ax4.set_ylabel('im $Z(f)$', color='r') >>> _ = ax3.set_xlabel('Frequency $f$ [Hz]') >>> _ = ax3.set_ylim(1e-2, 1) >>> _ = ax4.set_ylim(-0.2, 0) >>> pg.plt.show()
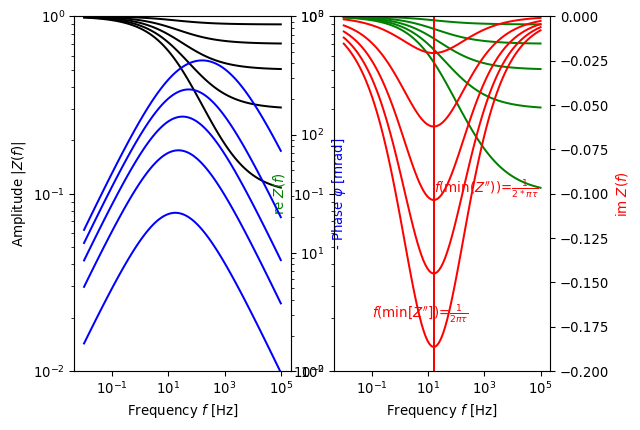
Examples using pygimli.physics.SIP.modelColeColeRho
- pygimli.physics.SIP.modelColeColeRhoDouble(f, rho, m1, t1, c1, m2, t2, c2, a=1, mult=False)[source]#
Frequency-domain double Cole-Cole resistivity (impedance) model.
Frequency-domain Double Cole-Cole impedance model returns the sum of two Cole-Cole Models with a common amplitude. Z = rho * (Z1(Cole-Cole) + Z2(Cole-Cole))
- pygimli.physics.SIP.modelColeColeSigma(f, sigma, m, tau, c, a=1)[source]#
Complex-valued conductivity (admittance) Cole-Cole model.
- pygimli.physics.SIP.modelColeColeSigmaDouble(f, sigma, m1, t1, c1, m2, t2, c2, a=1, tauRho=True)[source]#
Complex-valued double added conductivity (admittance) model.
- pygimli.physics.SIP.showSpectrum(freq, amp, phi, nrows=2, ylog=None, axs=None, **kwargs)[source]#
Show amplitude and phase spectra in two subplots.
- pygimli.physics.SIP.tauRhoToTauSigma(tRho, m, c)[source]#
Convert \(\tau_{\rho}\) to \(\tau_{\sigma}\) Cole-Cole model.
\[\tau_{\sigma} = \tau_{\rho}/(1-m)^{\frac{1}{c}}\]Examples
>>> import numpy as np >>> import pygimli as pg >>> from pygimli.physics.SIP import modelColeColeRho, modelColeColeSigma >>> from pygimli.physics.SIP import tauRhoToTauSigma >>> tr = 1. >>> Z = modelColeColeRho(1e5, rho=10.0, m=0.5, tau=tr, c=0.5) >>> ts = tauRhoToTauSigma(tr, m=0.5, c=0.5) >>> S = modelColeColeSigma(1e5, sigma=0.1, m=0.5, tau=ts, c=0.5) >>> abs(1.0/S - Z) < 1e-12 True >>> float(np.angle(1.0/S / Z)) < 1e-12 True
Classes#
- class pygimli.physics.SIP.ColeColeAbs(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Cole-Cole model with EM term after Pelton et al. (1978).
- __init__((object)arg1[, (object)verbose=False]) object :[source]#
- C++ signature :
void* __init__(_object* [,bool=False])
__init__( (object)arg1, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::DataContainer {lvalue} [,bool=False])
__init__( (object)arg1, (object)mesh [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh [,bool=False])
__init__( (object)arg1, (object)mesh, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh,GIMLI::DataContainer {lvalue} [,bool=False])
- class pygimli.physics.SIP.ColeColeComplex(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Cole-Cole model with EM term after Pelton et al. (1978).
- __init__((object)arg1[, (object)verbose=False]) object :[source]#
- C++ signature :
void* __init__(_object* [,bool=False])
__init__( (object)arg1, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::DataContainer {lvalue} [,bool=False])
__init__( (object)arg1, (object)mesh [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh [,bool=False])
__init__( (object)arg1, (object)mesh, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh,GIMLI::DataContainer {lvalue} [,bool=False])
- class pygimli.physics.SIP.ColeColeComplexSigma(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Cole-Cole model with EM term after Pelton et al. (1978).
- __init__((object)arg1[, (object)verbose=False]) object :[source]#
- C++ signature :
void* __init__(_object* [,bool=False])
__init__( (object)arg1, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::DataContainer {lvalue} [,bool=False])
__init__( (object)arg1, (object)mesh [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh [,bool=False])
__init__( (object)arg1, (object)mesh, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh,GIMLI::DataContainer {lvalue} [,bool=False])
- class pygimli.physics.SIP.ColeColePhi(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Cole-Cole model with EM term after Pelton et al. (1978).
Modelling operator for the Frequency Domain
Cole-Coleimpedance model usingpygimli.physics.SIP.modelColeColeRhoafter Pelton et al. (1978) [Pelton et al., 1978]\(\textbf{m} =\{ m, \tau, c\}\)
Modelling parameter for the Cole-Cole model with \(\rho_0 = 1\)
\(\textbf{d} =\{\varphi_i(f_i)\}\)
Modelling response for all given frequencies as negative phase angles \(\varphi(f) = -tan^{-1}\frac{\text{Im}\,Z(f)}{\text{Re}\,Z(f)}\) and \(Z(f, \rho_0=1, m, \tau, c) =\) Cole-Cole impedance.
- class pygimli.physics.SIP.DebyeComplex(fvec, tvec, verbose=False)[source]#
Bases:
ModellingDebye decomposition (smooth Debye relaxations) of complex data.
- class pygimli.physics.SIP.DebyePhi(fvec, tvec, verbose=False)[source]#
Bases:
ModellingDebye decomposition (smooth Debye relaxations) phase only.
- class pygimli.physics.SIP.DoubleColeColePhi(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Double Cole-Cole model after Pelton et al. (1978).
Modelling operator for the Frequency Domain - phase only
Cole-Coleimpedance model usingpygimli.physics.SIP.modelColeColeRhoafter Pelton et al. (1978) [Pelton et al., 1978]\(\textbf{m} =\{ m_1, \tau_1, c_1, m_2, \tau_2, c_2\}\)
Modelling parameter for the Cole-Cole model with \(\rho_0 = 1\)
\(\textbf{d} =\{\varphi_i(f_i)\}\)
Modelling Response for all given frequencies as negative phase angles \(\varphi(f) = \varphi_1(Z_1(f))+\varphi_2(Z_2(f)) = -tan^{-1}\frac{\text{Im}\,(Z_1Z_2)}{\text{Re}\,(Z_1Z_2)}\) and \(Z_1(f, \rho_0=1, m_1, \tau_1, c_1)\) and \(Z_2(f, \rho_0=1, m_2, \tau_2, c_2)\) ColeCole impedances.
- class pygimli.physics.SIP.PeltonPhiEM(f, verbose=False)[source]#
Bases:
ModellingBaseMT__Cole-Cole model with EM term after Pelton et al. (1978).
- __init__((object)arg1[, (object)verbose=False]) object :[source]#
- C++ signature :
void* __init__(_object* [,bool=False])
__init__( (object)arg1, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::DataContainer {lvalue} [,bool=False])
__init__( (object)arg1, (object)mesh [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh [,bool=False])
__init__( (object)arg1, (object)mesh, (object)dataContainer [, (object)verbose=False]) -> object :
- C++ signature :
void* __init__(_object*,GIMLI::Mesh,GIMLI::DataContainer {lvalue} [,bool=False])
- class pygimli.physics.SIP.SIPSpectrum(filename=None, unify=False, onlydown=True, f=None, amp=None, phi=None, k=1, sort=True, basename='new', **kwargs)[source]#
Bases:
objectSIP spectrum data analysis.
- __init__(filename=None, unify=False, onlydown=True, f=None, amp=None, phi=None, k=1, sort=True, basename='new', **kwargs)[source]#
Init SIP class with either filename to read or data vectors.
Examples
>>> #sip = SIPSpectrum('sipexample.txt', unify=True) # unique f values >>> #sip = SIPSpectrum(f=f, amp=R, phi=phase, basename='new')
- checkCRKK(useEps=False, use0=False, ax=None)[source]#
Check coupling removal (CR) by Kramers-Kronig (KK) relation.
- determineEpsilon(mode=0, sigmaR=None, sigmaI=None)[source]#
Retrieve frequency-independent epsilon for f->Inf.
- Parameters:
- Returns:
er – relative permittivity (epsilon) value (dimensionless)
- Return type:
- fit2CCPhi(ePhi=0.001, lam=1000.0, mpar=(0, 0, 1), taupar1=(0, 1e-05, 1), taupar2=(0, 0.1, 1000), cpar=(0.5, 0, 1), verbose=False)[source]#
Fit two Cole-Cole terms (to phase only).
- Parameters:
ePhi (float) – absolute error of phase angle
lam (float) – regularization parameter
mpar (list[3]) – starting value, lower bound, upper bound for chargeability
taupar2 (taupar1 /) – starting value, lower bound, upper bound for 2 time constants
cpar2 (cpar1 /) – starting value, lower bound, upper bound for 2 relaxation exponents
- fitCCEM(ePhi=0.001, lam=1000.0, remove=True, mpar=(0.2, 0, 1), taupar=(0.01, 1e-05, 100), cpar=(0.25, 0, 1), empar=(1e-07, 1e-09, 1e-05), verbose=False)[source]#
Fit a Cole-Cole term with additional EM term to phase.
- Parameters:
ePhi (float) – absolute error of phase angle
lam (float) – regularization parameter
remove (bool) – remove EM term from data
mpar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
taupar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
cpar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
empar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
- fitCCPhi(ePhi=0.001, lam=1000.0, mpar=(0, 0, 1), taupar=(0, 1e-05, 100), cpar=(0.3, 0, 1), verbose=False)[source]#
Fit a Cole-Cole term (to phase only).
- Parameters:
ePhi (float) – absolute error of phase angle
lam (float) – regularization parameter
mpar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
taupar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
cpar (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c) and EM relaxation time (em)
- fitColeCole(useCond=False, **kwargs)[source]#
Fit a Cole-Cole model to the phase data.
- Parameters:
useCond (bool) – use conductivity form of Cole-Cole model instead of resistivity
error (float [0.01]) – absolute phase error
lam (float [1000]) – initial regularization parameter
mpar (tuple/list (0, 0, 1)) – inversion parameters for chargeability: start, lower, upper bound
taupar (tuple/list (1e-2, 1e-5, 100)) – inversion parameters for time constant: start, lower, upper bound
cpar (tuple/list (0.25, 0, 1)) – inversion parameters for Cole exponent: start, lower, upper bound
- fitDebyeModel(ePhi=0.001, lam=1000.0, lamFactor=0.8, tau=None, mint=None, maxt=None, nt=None, useComplex=True, showFit=False, verbose=False, **kwargs)[source]#
Fit a (smooth) continuous Debye model (Debye decomposition).
- Parameters:
ePhi (float) – absolute error of phase angle
lam (float) – regularization parameter
lamFactor (float) – regularization factor for subsequent iterations
mint/maxt (float) – minimum/maximum tau values to use (else automatically from f)
nt (int) – number of tau values (default number of frequencies * 2)
new (bool) – new implementation (experimental)
showFit (bool) – show fit
cType (int) – constraint type (1/2=smoothness 1st/2nd order, 0=minimum norm)
phi (iterable) – use phi instead of self.phi
- fitDoubleColeCole(ePhi=0.001, eAmp=0.01, lam=1000.0, robust=False, verbose=True, useRho=True, useMult=False, aphi=True, mpar1=(0.2, 0, 1), mpar2=(0.2, 0, 1), tauRho=True, taupar1=(0.01, 1e-05, 100), taupar2=(0.0001, 1e-05, 100), cpar1=(0.5, 0, 1), cpar2=(0.5, 0, 1))[source]#
Fit double Cole-Cole term to complex resistivity or phase.
- Parameters:
ePhi (float [0.001]) – absolute error of phase angle in rad
eAmp (float [0.01 = 1%]) – absolute error of phase angle
lam (float) – regularization parameter
robust (bool [False]) – use robustData (L1 norm on data side)
useRho (bool [True]) – Cole-Cole defined for impedance/resistivity, otherwise conductivity
useMult (bool [False]) – the two terms are combined as product (otherwise sum)
tauRho (bool [False]) – in case of useRho=False the time constant is defined like for rho
mpar1/2 (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c)
taupar1/2 (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c)
cpar1/2 (list[3]) – inversion parameters (starting value, lower bound, upper bound) for Cole-Cole parameters (m, tau, c)
- loadData(filename, **kwargs)[source]#
Import spectral data.
Import Data and try to assume the file format.
- removeEpsilonEffect(er=None, mode=0)[source]#
Remove effect of (constant high-frequency) epsilon from sigma.
- showData(reim=False, znorm=False, cond=False, nrows=2, ax=None, **kwargs)[source]#
Show amplitude and phase spectrum in two subplots.
- zNorm()[source]#
Normalized real (difference) and imag. z [Nordsiek and Weller, 2008].
- class pygimli.physics.SIP.SpectrumManager(fop=None, **kwargs)[source]#
Bases:
MethodManagerManager to work with spectra data.
- __init__(fop=None, **kwargs)[source]#
Set up spectrum manager.
- Parameters:
fop (pg.Modelling operator, optional) – Forward operator. The default is None.
**kwargs (TYPE) – passed to SpectrumManager.
- class pygimli.physics.SIP.SpectrumModelling(funct=None, **kwargs)[source]#
Bases:
ParameterModellingModelling framework with an array of freqencies as data space.
- __init__(funct=None, **kwargs)[source]#
Initialize.
- Parameters:
func (function) – modelling function
complex (bool) – complex function
frequencies (iterable) – frequency vector
- property complex#
Return if spectrum is complex.
- property freqs#
Return frequency vector.