pygimli.physics.gravimetry#
Solve gravimetric and magneto static problems in 2D and 3D analytically
Overview#
Functions
|
Vertical magnetic gradient for polygone. |
|
Magnetic anomaly for a horizontal cylinder. |
|
Magnetic anomaly for a sphere. |
|
|
|
TODO WRITEME. |
|
TODO WRITEME. |
|
TODO WRITEME. |
|
2D Gradient of gravimetric potential of horizontal cylinder. |
|
Gravitational field od a horizontal half plate. |
|
Gravitational field of a sphere. |
|
Solve gravimetric response. |
|
Gravitational potential of horizonzal cylinder. |
|
Gravitational potential of a sphere. |
Classes
|
Magnetics modelling operator using Holstein (2007). |
|
Gravimetry modelling operator. |
|
Magnetics Manager. |
|
Magnetics modelling operator using Holstein (2007). |
Functions#
- pygimli.physics.gravimetry.BZPoly(pnts, poly, mag, openPoly=False)[source]#
Vertical magnetic gradient for polygone.
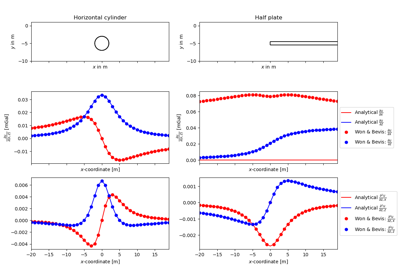
- pygimli.physics.gravimetry.BaZCylinderHoriz(pnts, R, pos, M)[source]#
Magnetic anomaly for a horizontal cylinder.
Calculate the vertical component of the anomalous magnetic field Bz for a buried horizontal cylinder at position pos with radius R for a given magnetization M at measurement points pnts.
TODO .. only 2D atm
- pygimli.physics.gravimetry.BaZSphere(pnts, R, pos, M)[source]#
Magnetic anomaly for a sphere.
Calculate the vertical component of the anomalous magnetic field Bz for a buried sphere at position pos with radius R for a given magnetization M at measurement points pnts.
- pygimli.physics.gravimetry.SolveGravMagHolstein(*args, **kwargs)#
- pygimli.physics.gravimetry.gradGZCylinderHoriz(r, a, rho, pos=(0.0, 0.0))[source]#
TODO WRITEME.
\[g = -grad u(r), with r = [x,z], |r| = \sqrt{x^2+z^2}\]
Examples using pygimli.physics.gravimetry.gradGZCylinderHoriz
- pygimli.physics.gravimetry.gradGZHalfPlateHoriz(pnts, t, rho, pos=(0.0, 0.0))[source]#
TODO WRITEME.
\[g = -\nabla u\]
Examples using pygimli.physics.gravimetry.gradGZHalfPlateHoriz
- pygimli.physics.gravimetry.gradGZSphere(r, rad, rho, pos=(0.0, 0.0, 0.0))[source]#
TODO WRITEME.
\[g = -\nabla u\]
- pygimli.physics.gravimetry.gradUCylinderHoriz(r, a, rho, pos=(0.0, 0.0))[source]#
2D Gradient of gravimetric potential of horizontal cylinder.
\[g = -G[m^3/(kg s^2)] * dM[kg/m] * 1/r[1/m] * grad(r)[1/1] = [m^3/(kg s^2)] * [kg/m] * 1/m * [1/1] == m/s^2\]
Examples using pygimli.physics.gravimetry.gradUCylinderHoriz
- pygimli.physics.gravimetry.gradUHalfPlateHoriz(pnts, t, rho, pos=(0.0, 0.0))[source]#
Gravitational field od a horizontal half plate.
\[g = -grad u,\]- Parameters:
pnts
t
rho – Density in [kg/m^3]
- Returns:
z-component of g .. math:: nabla(partial u/partialvec{r})_z
- Return type:
gz
Examples using pygimli.physics.gravimetry.gradUHalfPlateHoriz
- pygimli.physics.gravimetry.gradUSphere(r, rad, rho, pos=(0.0, 0.0, 0.0))[source]#
Gravitational field of a sphere.
\[g = -G[m^3/(kg s^2)] * dM[kg] * 1/r^2 1/m^2] * \grad(r)[1/1] = [m^3/(kg s^2)] * [kg] * 1/m^2 * [1/1] == m/s^2\]
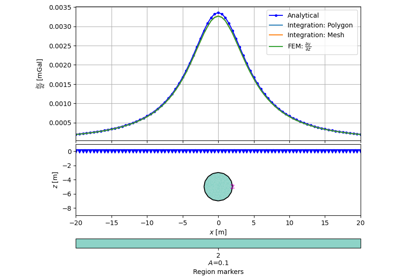
- pygimli.physics.gravimetry.solveGravimetry(mesh, dDensity=None, pnts=None, complete=False)[source]#
Solve gravimetric response.
2D with
pygimli.physics.gravimetry.lineIntegralZ_WonBevis3D with
pygimli.physics.gravimetry.gravMagBoundarySinghGup- Parameters:
mesh (GIMLI::Mesh) – 2d or 3d mesh with or without cells.
dDensity (float | array) –
Density difference.
- float – solve for positive boundary marker only.
Assuming one inhomogeneity.
[[int, float]] – solve for multiple positive boundaries TOIMPL
array – solve for one delta density value per cell
None – return per cell kernel matrix G TOIMPL
pnts ([[x_i, y_i]]) – List of measurement positions.
complete (bool [False]) – If True return whole solution or matrix for [dgx, dgy, dgz]
- Returns:
dg (array OR)
dz, dgz (arrays (if complete))
Examples using pygimli.physics.gravimetry.solveGravimetry
- pygimli.physics.gravimetry.uCylinderHoriz(pnts, rad, rho, pos=[0.0, 0.0])[source]#
Gravitational potential of horizonzal cylinder.
Classes#
- class pygimli.physics.gravimetry.GravityModelling(mesh, points, cmp=['gz'])[source]#
Bases:
MeshModellingMagnetics modelling operator using Holstein (2007).
- class pygimli.physics.gravimetry.GravityModelling2D(points=None, **kwargs)[source]#
Bases:
MeshModellingGravimetry modelling operator.
- __init__(points=None, **kwargs)[source]#
Initialize forward operator, optional with mesh and points.
You can specify both the mesh and the measuring points, or set the latter after the mesh has been set.
- Parameters:
mesh (pg.Mesh) – mesh for forward computation
points (array[x,y]) – measuring points
- class pygimli.physics.gravimetry.MagManager(data=None, **kwargs)[source]#
Bases:
MeshMethodManagerMagnetics Manager.
- createMesh(bnd=0, area=100000.0, depth=800, quality=1.3, addPLC=None, addPoints=True)[source]#
Create an unstructured mesh.
- inversion(noise_level=2, noisify=False, **kwargs)[source]#
Run Inversion (requires mesh and FOP).
- Parameters:
noise_level (float|array) – absolute noise level (absoluteError)
noisify (bool) – add noise before inversion
relativeError (float|array [0.01]) – relative error to stabilize very low data
depthWeighting (bool [True]) – apply depth weighting after Li&Oldenburg (1996)
z0 (float) – skin depth for depth weighting
mul (array) – multiply constraint weight with
arguments (standard inversion keyword)
....................................
C( (int|Matrix|[float, float, float]) – constraint order, matrix or correlation lengths
cType) (int|Matrix|[float, float, float]) – constraint order, matrix or correlation lengths
symlogThreshold (float [0]) – threshold for symlog data trans (0 = linear)
startModel (float|array) – starting model (typically homogeneous)
lam (float) – regularization strength
robustData (bool) – L1 norm on data misfit and model roughness
blockyModel (bool) – L1 norm on data misfit and model roughness
maxIter (int) – maximum iteration number
- Returns:
model – model vector (also saved in self.inv.model)
- Return type:
array
- class pygimli.physics.gravimetry.MagneticsModelling(mesh=None, points=None, cmp=['TFA'], igrf=[50, 13])[source]#
Bases:
MeshModellingMagnetics modelling operator using Holstein (2007).
- __init__(mesh=None, points=None, cmp=['TFA'], igrf=[50, 13])[source]#
Setup forward operator.
- Parameters:
mesh (pygimli:mesh) – tetrahedral or hexahedral mesh
points (list|array of (x, y, z)) – measuring points
cmp (list of str) – component of: gx, gy, gz, TFA, Bx, By, Bz, Bxy, Bxz, Byy, Byz, Bzz
igrf (list|array of size 3 or 7) –
international geomagnetic reference field, either [D, I, H, X, Y, Z, F] - declination, inclination, horizontal field,
X/Y/Z components, total field OR
[X, Y, Z] - X/Y/Z components [lat, lon] - latitude, longitude (automatic IGRF)