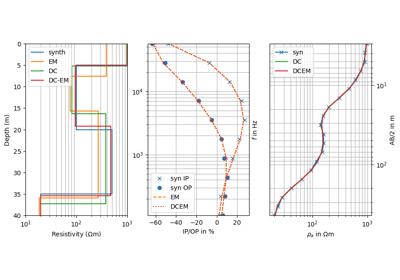
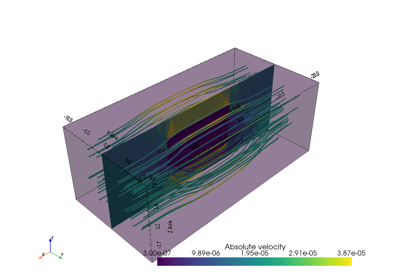
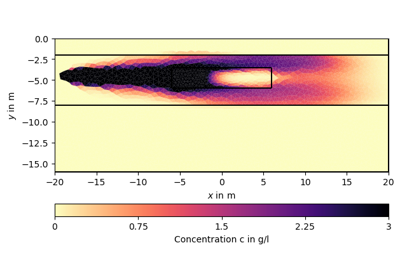
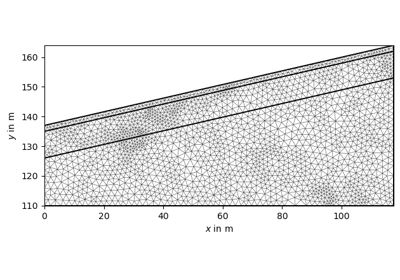
import numpy as np
import pygimli as pg
from .utils import pgMesh2pvMesh
pv = pg.optImport('pyvista', requiredFor="properly visualize 3D data")
[docs]def drawMesh(ax, mesh, notebook=False, **kwargs):
"""
Parameters
----------
ax: pyvista.Plotter [optional]
The plotter to draw everything. If none is given, one will be created.
mesh: pg.Mesh
The mesh to show.
notebook: bool [False]
Sets the plotter up for jupyter notebook/lab.
cmap: str ['viridis']
The colormap string.
Returns
-------
ax: pyvista.Plotter [optional]
The plotter
"""
# sort out a few kwargs to not confuse the plotter initialization
show_edges = kwargs.pop('show_edges', True)
opacity = kwargs.pop('alpha', kwargs.pop('opacity', 1))
cmap = kwargs.pop('cmap', None)
color = kwargs.pop('color', 'k')
style = kwargs.pop('style', 'wireframe')
returnActor = kwargs.pop('returnActor', False)
if ax is None:
# if notebook:
# ax = pv.PlotterITK(**kwargs)
# else:
ax = pv.Plotter(notebook=notebook, **kwargs)
ax.show_bounds(all_edges=True, minor_ticks=True)
ax.add_axes()
if isinstance(mesh, pg.Mesh):
mesh = pgMesh2pvMesh(mesh)
_actor = ax.add_mesh(mesh, # type: pv.UnstructuredGrid
cmap=cmap,
color=color,
style=style,
show_edges=show_edges,
opacity=opacity,
)
if returnActor:
return ax, _actor
else:
return ax
[docs]def drawModel(ax=None, mesh=None, data=None, **kwargs):
"""
Draw the mesh with given data.
Parameters
----------
ax: pyvista.Plotter [None]
Pyvista's basic Plotter to add the mesh to.
mesh: pg.Mesh
The Mesh to plot.
data: iterable
Data that should be displayed with the mesh.
Returns
-------
ax: pyvista.Plotter [optional]
The plotter
"""
if all(v is None for v in [ax, mesh, data]):
pg.critical("At least mesh or data should not be None")
return None
if data is not None or len(mesh.dataMap()) != 0:
kwargs['style'] = 'surface'
kwargs['color'] = None
mesh = pgMesh2pvMesh(mesh, data, kwargs.pop('label', None))
if 'cmap' not in kwargs:
kwargs['cmap'] = 'viridis'
return drawMesh(ax, mesh, **kwargs)
[docs]def drawSensors(ax, sensors, diam=0.01, color='grey', **kwargs):
"""
Draw the sensor positions to given mesh or the the one in given plotter.
Parameters
----------
ax: pyvista.Plotter
The plotter to draw everything. If none is given, one will be created.
sensors: iterable
Array-like object containing tuple-like (x, y, z) positions.
diam: float [0.01]
Radius of sphere markers.
color: str ['grey']
Color of sphere markers.
Returns
-------
ax: pyvista.Plotter
The plotter containing the mesh and drawn electrode positions.
"""
for pos in sensors:
s = pv.Sphere(radius=diam / 2, center=pos)
ax.add_mesh(s, color=color, **kwargs)
return ax
[docs]def drawSlice(ax, mesh, normal=[1, 0, 0], **kwargs):
"""
Parameters
----------
ax: pyvista.Plotter
The Plotter to draw on.
mesh: pg.Mesh
The mesh to take the slice out of.
normal: list [[1, 0, 0]]
Coordinates to orientate the slice.
Returns
-------
ax: pyvista.Plotter
The plotter containing the mesh and drawn electrode positions.
Note
----
Possible kwargs are:
normal: tuple(float), str
origin: tuple(float)
generate_triangles: bool, optional
contour: bool, optional
They can be found at https://docs.pyvista.org/core/filters.html#pyvista.CompositeFilters.slice
"""
label = kwargs.pop('label', None)
data = kwargs.pop('data', None)
mesh = pgMesh2pvMesh(mesh, data, label)
try:
single_slice = mesh.slice(normal, **kwargs)
except AssertionError as e:
# 'contour' kwarg only works with point data and breaks execution
pg.error(e)
else:
# REVIEW: bounds and axes might be confused with the outline..?!
outline = mesh.outline()
ax.add_mesh(outline, color="k")
ax.add_mesh(single_slice)
return ax
[docs]def drawStreamLines(ax, mesh, data, label=None, radius=0.01, **kwargs):
"""
Draw streamlines of given data.
PyVista streamline needs a vector field of gradient data per cell.
Parameters
----------
ax: pyvista.Plotter [None]
The plotter that should be used for visualization.
mesh: pyvista.UnstructuredGrid|pg.Mesh [None]
Structure to plot the streamlines in to.
If its a pv grid a check is performed if the data set is already contained.
data: iterable [None]
Values used for streamlining.
label: str
Label for the data set. Will be searched for within the data.
radius: float [0.01]
Radius for the streamline tubes.
Note
----
All kwargs will be forwarded to pyvistas streamline filter:
https://docs.pyvista.org/core/filters.html?highlight=streamlines#pyvista.DataSetFilters.streamlines
"""
if isinstance(mesh, pg.Mesh):
# create gradient of cell data if not provided
if np.ndim(data) == 1:
grad = pg.solver.grad(mesh, data)
else:
grad = data
# ensure that it's point/node data in the mesh
if len(grad) == mesh.cellCount():
grad = pg.meshtools.cellDataToNodeData(mesh, grad)
# add data to the mesh and convert to pyvista grid
mesh = pgMesh2pvMesh(mesh, grad.T, label)
elif isinstance(mesh, pv.UnstructuredGrid):
if label not in mesh.point_arrays: # conversion needed
mesh.cell_data_to_point_data()
if label is None:
label = list(mesh.point_arrays.keys())[0]
kwargs['vectors'] = label
streamlines = mesh.streamlines(**kwargs)
ax.add_mesh(streamlines.tube(radius=radius))